RG2: Molecular Biomarkers and Diagnosis
This cross-disciplinary RG focused on translational research on diseases spanning the three Thematic Lines, from cancer to neurodegeneration and to inflammatory diseases & diabetes, revolves around three main axes:
- Identification of potential biomarkers (BM) and therapeutic targets (employing experimental approaches & computation data integration and modelling);
- Development of antibodies (Abs) targeting the molecules identified;
- Establishment and miniaturization of new assays.
BM identification for diagnostics and therapy design leverage precision medicine approaches. Our activities include discovery of BM for diagnosis, patient stratification & therapy design. Complementary, we develop Abs for BM detection and as analytical tools for pre- and clinical settings. Our activities are leveraged by the group expertise on glyco-, neuro- and onco-biology, cellular biotechnology, microfluidics and computational modelling, strongly complemented by state-of-the-art platforms established in-house: proteomics, glycomics, disease cell modelling and Ab discovery. These systematic technological approaches combined with data integration are fundamental for BM discovery.
Specifically, for the identification/quantification of deregulated proteins and their post-translational modifications in disease, we employ patient samples/patient-derived cell models developed within the group. We focus on cell content and extracellular vesicles, important targets for disease BM identification. The samples are interrogated using a wide range of techniques, eg., MS, NMR, HPLC, NGS, cell imaging; the large-scale molecular datasets generated are analysed by advanced computational models. We translate the generated knowledge to the development of novel therapeutic Abs/establishment of new assays. Recently, we established a high-throughput microfluidic platform for identification of novel Abs, based on single cell analysis of Ab-producing B cells.
The main objective of RG2 is identifying BM and developing tools to facilitate the translation of the knowledge to the clinical groups within iNOVA4Health. Within this context, we highlight 3 activities:
1. Discovery of disease biomarkers
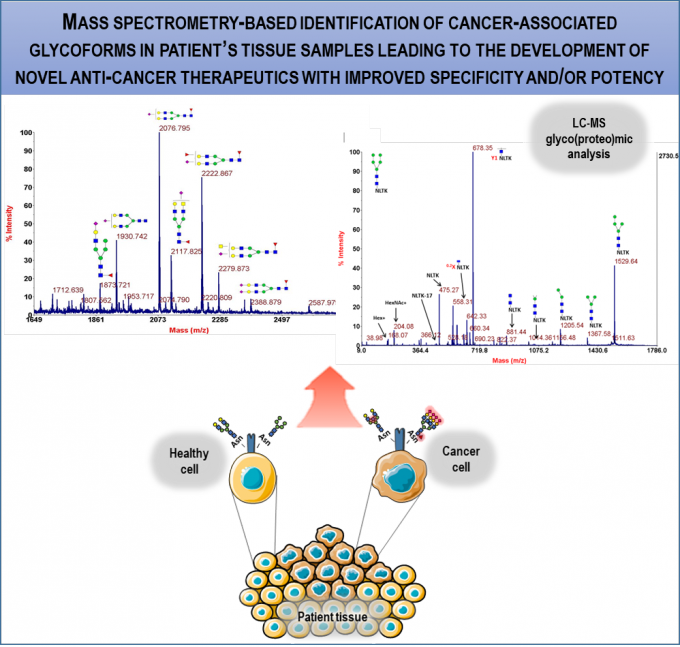
RG2 has been working on the identification of disease BM for: i) cancer; ii) amyotrophic lateral sclerosis - ALS; axial apondyloarthritis - AS. For identification of cancer BM, we implemented MS-based glycoprote- & glyc-omics approaches for characterization of glycosignatures of disease vs normal biological samples. We have glycoprofiled extracellular vesicles from ovarian carcinoma and glioma cell lines and specific cell surface receptors in samples from patients with head & neck and oesophageal carcinomas (within TL1, collaboration with Merck). MS analysis of bronchoalveolar lavage (BAL) samples revealed remarkable overlap with markers of lung cancer tissue compared to patient-matched normal tissue; extracellular vesicles from BAL are being studied as potential BM. In the context of an EC-funded consortium, we identified phosphoneurofilament heavy chain and explored additional molecules as ALS BM. In AS, we are correlating protein & glycan signatures with clinical data on patient muscle properties along disease progression to identify potential BM for diagnosis and preventive/therapeutic approaches (within TL2).
2. Patient-derived cancer cell model platform
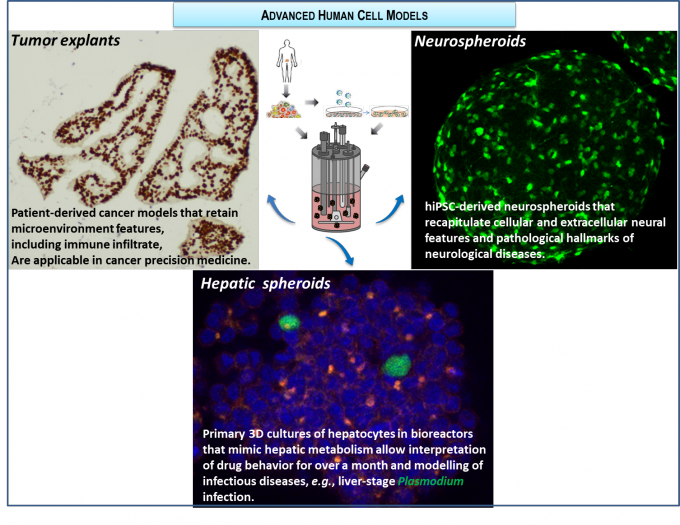
RG2 has been developing ex vivo cancer model platforms, recapitulating the tumour microenvironment and compatible with cyclic exposure to therapeutic compounds. Firstly, we established a proof-of-concept with cancer cell lines combined with fibroblasts and lately including immune cells, demonstrating recapitulation of phenotypic features of disease progression induced by tumor, fibroblast and macrophage crosstalk. Recently, we have developed methodologies for long-term culture of colorectal, ovary and breast cancer patient-derived samples; more than 60 explants derived from patient tumor material were successfully cultured, retaining the original tissue architecture and main cellular components; we are currently evaluating standard of care and new therapeutic avenues, towards a precision medicine platform (within TL3, in collaboration with Abbvie and IPOLFG).
3. Phage display-based platform for Ab discovery
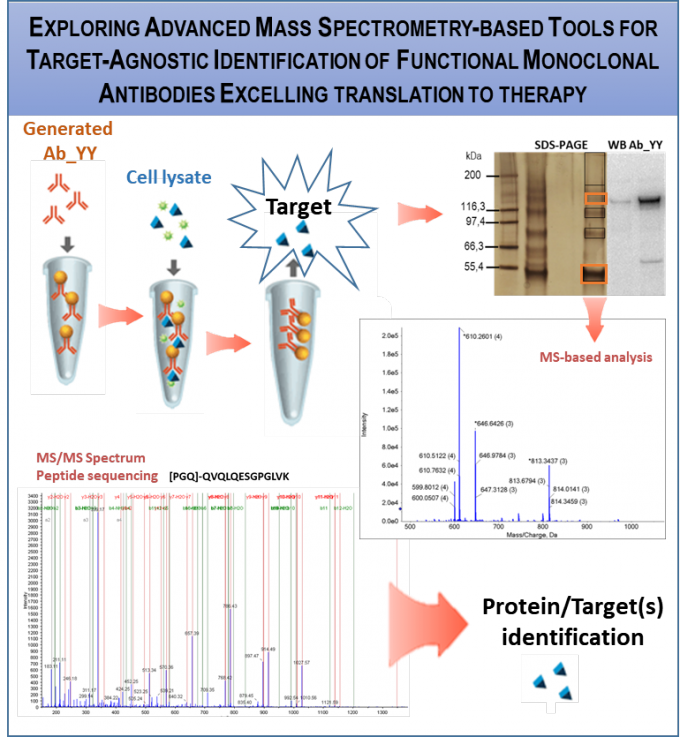
RG2 is generating phage display libraries for identification of human Abs that bind/regulate the function of target proteins. We developed 2 libraries, employing breast cancer patient and healthy donor samples and applied them to discover function blocking Abs targeting Notch signalling that present anti-oncogenic activity in breast cancer. We are expanding library application to other diseases, eg., type 2 diabetes (T2D), by developing Ab-binding assays for the identification of gut-derived exosomes, recognized as potential T2D biomarkers.(collaborative projects within TL2 and 3).
Keywords: Proteomics; Glycomics; Antibodies; Cancer
Latest Publications
Erny GL, Gomes RA, Santos MSF, Santos L, Neuparth N, Carreiro-Martins P, Marques JG, Guerreiro ACL, Gomes-Alves P (2020) Mining for Peaks in LC-HRMS Datasets Using Finnee – A Case Study with Exhaled Breath Condensates from Healthy, Asthmatic, and COPD Patients. ACS Omega 5(26): 16089-16098
Lopes-Coelho F, Silva F, Gouveia-Fernandes S, Martins C, Lopes N, Domingues G, Brito C, Almeida AM, Pereira SA, Serpa J (2020) Monocytes as endothelial percursor cells (EPCs), another brick in the wall to disentangle tumor angiogenesis. Cells. 9(1):107
Sales-Dias J, Silva G, Lamy M, Ferreira A, Barbas A (2019) The Notch ligand DLL1 exerts carcinogenic features in human breast cancer cells. PLoS One 14(5):e0217002
Serra AT, Serra M, Silva AC, Brckalo T, Seshire A, Brito C, Wolf M, Alves PM (2019) Scalable culture strategies for the expansion of patient-derived cancer stem cell lines. Stem Cells International Article ID 8347595
Lima RT, Sousa D, Gomes AS, Mendes N, Matthiesen R, Pedro M, Marques F, Pinto MM, Sousa E, Vasconcelos MH (2019) The antitumor activity of a lead thioxanthone is associated with alterations in cholesterol localization. Molecules 23(12):3301